This page was generated from docs/source/examples/GroupedPipeline.ipynb.
GroupedPipeline: applying a transformer per category¶
[1]:
import numpy as np
import pandas as pd
from sklearn.datasets import load_iris
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error as mse, mean_absolute_error as mae
from timeserio.data.datasets import load_iris_df
from timeserio.pipeline import GroupedPipeline
from timeserio.preprocessing import PandasValueSelector
import seaborn as sns
Load the iris dataset¶
This dataset consists of four numeric and one categorical columns.
[2]:
df = load_iris_df()
df.head(2)
[2]:
| sepal_length_cm | sepal_width_cm | petal_length_cm | petal_width_cm | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
Imagine we want to predict sepal_width_cm based on sepal_length_cm: we may try fitting a simple linear regression model. The result is somewhat unsatisfying:
Fit a joint model¶
[3]:
sns.lmplot(x="sepal_length_cm", y="sepal_width_cm", data=df)
[3]:
<seaborn.axisgrid.FacetGrid at 0x7f600c6d2ba8>
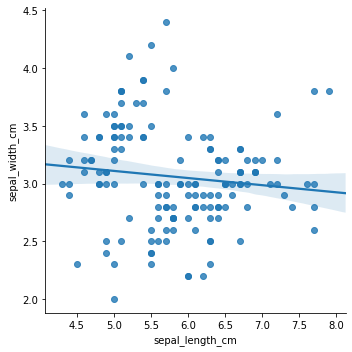
In sklearn, we can represent the model (in this case, a linear regression on selected features) as a pipeline:
[4]:
simple_model = Pipeline([
("features", PandasValueSelector("sepal_length_cm")),
("lr", LinearRegression()),
])
[5]:
simple_model.fit(df, df["sepal_width_cm"])
[5]:
Pipeline(memory=None,
steps=[('features', PandasValueSelector(columns='sepal_length_cm')), ('lr', LinearRegression(copy_X=True, fit_intercept=True, n_jobs=None,
normalize=False))])
[6]:
y_pred, y = simple_model.predict(df), df["sepal_width_cm"]
print(f"MSE = {mse(y, y_pred)}, MAE = {mae(y, y_pred)}")
MSE = 0.18610437589381357, MAE = 0.33122352979559877
[7]:
lr = simple_model.named_steps['lr']
print(f"Intercept: {lr.intercept_:.2f}, slope: {lr.coef_[0]:.2f}")
Intercept: 3.42, slope: -0.06
Fit a model per category¶
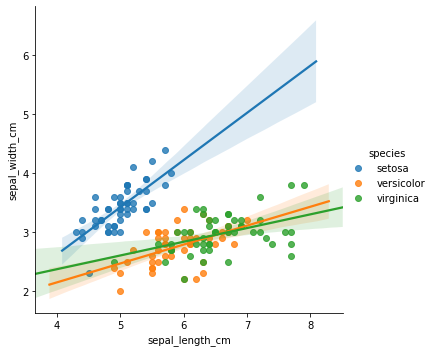
We can improve the regression model signiifcantly by adding the categorical feature species - we could try on-hot encoding, embeddings, etc. However, a common approach would be to fit a separate regression model per category:
[8]:
sns.lmplot(x="sepal_length_cm", y="sepal_width_cm", hue="species", data=df)
[8]:
<seaborn.axisgrid.FacetGrid at 0x7f600bee2860>
[9]:
grouped_model = GroupedPipeline(
groupby="species",
pipeline=simple_model
)
[10]:
grouped_model.fit(df, df["sepal_width_cm"])
# Specifying target by column name is also supported:
grouped_model.fit(df, "sepal_width_cm")
[10]:
GroupedPipeline(errors='raise', groupby='species',
pipeline=Pipeline(memory=None,
steps=[('features', PandasValueSelector(columns='sepal_length_cm')), ('lr', LinearRegression(copy_X=True, fit_intercept=True, n_jobs=None,
normalize=False))]))
[11]:
y_pred, y = grouped_model.predict(df), df["sepal_width_cm"]
print(f"MSE = {mse(y, y_pred)}, MAE = {mae(y, y_pred)}")
MSE = 0.07119979049915942, MAE = 0.20655162210804365
[12]:
for group, pipe in grouped_model.pipelines_.items():
lr = pipe.named_steps['lr']
print(f"{group}: intercept: {lr.intercept_:.2f}, slope: {lr.coef_[0]:.2f}")
setosa: intercept: -0.57, slope: 0.80
versicolor: intercept: 0.87, slope: 0.32
virginica: intercept: 1.45, slope: 0.23